-
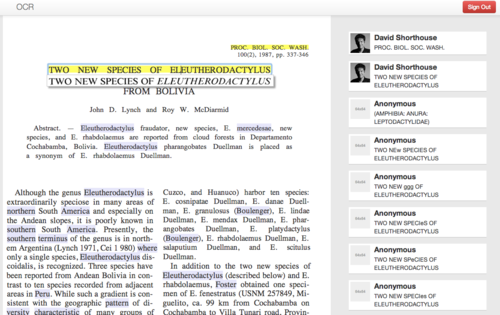
An in-place panel shows the exact line in the original scanned image while the user edits a single line of OCR text at a time (Figure 1)
-
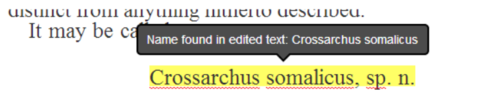
Global Names scientific name-finding is integrated in real-time when a user completes a line edit, giving feedback if a scientific name is newly recognized (Figure 2)
-
Authentication uses the facile https://oauth.io/ such that all edits are tied to users’ OAuth2-provider accounts (eg Google, Twitter, GitHub)
-
Frequencies of common edits are summarized in real-time and other words that may benefit from similar edits are highlighted for users
-
Batch processes collapse all user edits and text files are recreated for possible re-introduction into data mining routines
-
Unit and integration tests are included
The event wiki page here has been recently updated with additional information on the different series of activities organised (workshops, trainings and demonstrations) and the Final Conference agenda now comprises worldwide high-level speakers, including (i) officials from the European Commission DG Connect, the US National Academy of Sciences, (ii) representatives from botanic gardens, natural museums, other biodiversity initiatives and (iii) experts or (iv) researchers specialized in biodiversity informatics, environmental/natural science.
One of the key objectives of these series of events will be to ensure the Final event will provide key recommendations and inputs from biodiversity experts for the preparation of the next WP 2016-2017 as specifically asked by the European Commission.
The number of registered attendees has already reached a good level of participation to insure a thorough exchange of information and experience between stakeholders interested in making fundamental biodiversity data digital, open and re-usable. Visit the different activities pages to find out more on the attendance status.
In this context, if you plan to attend and have not yet registered, we can only recommend you do to it as soon as possible here (due to room capacity constraints).
For further information on this event (agenda, concept & objectives, registration) please visit the Event wiki page or contact us at final-event@pro-ibiosphere.eu.

 A group of scientists and students discovered the new species of spider during a field course in Borneo, supervised by Jeremy Miller and Menno Schilthuizen from the Naturalis Biodiversity Center, based in Leiden, the Netherlands. The species was described and submitted online from the field to the Biodiversity Data Journal through a satellite internet connection, along with the underlying data . The manuscript was peer-reviewed and published within two weeks of submission. On the day of publication, GBIF and EOL have harvested and included the data in their respective platforms.
A group of scientists and students discovered the new species of spider during a field course in Borneo, supervised by Jeremy Miller and Menno Schilthuizen from the Naturalis Biodiversity Center, based in Leiden, the Netherlands. The species was described and submitted online from the field to the Biodiversity Data Journal through a satellite internet connection, along with the underlying data . The manuscript was peer-reviewed and published within two weeks of submission. On the day of publication, GBIF and EOL have harvested and included the data in their respective platforms.
Workshop on Model Evaluation
Wednesday June 11 (all day)
Demonstrations on pro-iBiosphere pilots
Demonstrations on outcomes of pro-iBiosphere Data Enrichment Hackathon
Workshop on Biodiversity Catalogue
Training on WikiMedia
Poster session
Thursday June 12 (all day)
Final Conference
Networking Cocktail
 piB Final Event announcement
piB Final Event announcement
The iMarine initiative provides a data infrastructure aimed at facilitating open access, the sharing of data, collaborative analysis, processing and mining processing, as well as the dissemination of newly generated knowledge. The iMarine data infrastructure is developed to support decision making in high-level challenges that require policy decisions typical of the ecosystem approach.
iMarine has developed a series of applications which can be clustered in four main thematic domains (the so called Application Bundles, set of services and technologies grouped according to a family of related tasks for achieving a common objective).
More information on the iMarine Catalogue of Applications here.

BioVeL announces the upcoming release of the BioVeL Portal. Designed in response to scientists' needs through a continuous cycle of requests and feedback, the portal will be robust and scalable for handling greater workloads.
An important feature of the Portal will be the ability to do "data sweeps"– that is, to initiate multiple runs of the same workflow, each with different input conditions. Other neat points are the organisation of the workflows by categories with "facetted browsing" for easier search, and a complete history of all your own workflow runs. Also through the Portal you can share workflows and results between collaborators. And as always with BioVeL tools, the codebase used for the portal benefits from being used across multiple projects.
Access BioVeL Portal here.
Please do not hesitate to provide your comments to contact@biovel.eu

The pro-iBiosphere project organied 2 workshops between February 10-12 in Berlin:
- February 10: MS12 - Workshop on mark-up of biodiversity literature
- February 11: MS12 - Workshop on mark-up of biodiversity literature and MS23 - Workshop on alternative business models
- February 12: MS23 - Workshop on alternative business models
The workshops took place at the Museum für Naturkunde Berlin (MFN) located 43 Invalidenstraße in Berlin.
For complementary information on these events (concept, objectives and outcomes), please visit the dedicated project wiki page here or contact us: info@pro-ibiosphere.eu.

Today, 16/12/2013, the European Commission announced the launch of a new Pilot on Open Research Data in Horizon 2020, to ensure that valuable information produced by researchers in many EU-funded projects will be shared freely. Researchers in projects participating in the pilot are asked to make the underlying data needed to validate the results presented in scientific publications and other scientific information available for use by other researchers, innovative industries and citizens. This will lead to better and more efficient science and improved transparency for citizens and society. It will also contribute to economic growth through open innovation. For 2014-2015, topic areas participating in the Open Research Data Pilot will receive funding of around €3 billion.
The Commission recognises that research data is as important as publications. It therefore announced in 2012 that it would experiment with open access to research data (see IP/12/790). The Pilot on Open Research Data in Horizon 2020 does for scientific information what the Open Data Strategy does for public sector information: it aims to improve and maximise access to and re-use of research data generated by projects for the benefit of society and the economy.
The Pilot involves key areas of Horizon 2020:
-
Future and Emerging Technologies
-
Research infrastructures – part e-Infrastructures
-
Leadership in enabling and industrial technologies – Information and Communication Technologies
-
Societal Challenge: Secure, Clean and Efficient Energy – part Smart cities and communities
-
Societal Challenge: Climate Action, Environment, Resource Efficiency and Raw materials – with the exception of topics in the area of raw materials
-
Societal Challenge: Europe in a changing world – inclusive, innovative and reflective Societies
-
Science with and for Society
Neelie Kroes, Vice-President of the European Commission for the Digital Agenda said "We know that sharing and re-using research data holds huge potential for science, society and the economy. This Pilot is an opportunity to see how different disciplines share data in practice and to understand remaining obstacles."
Commissioner Máire Geoghegan-Quinn said: "This pilot is part of our commitment to openness in Horizon 2020. I look forward to seeing the first results, which will be used to help set the course for the future."
Projects may opt out of the pilot to allow for the protection of intellectual property or personal data; in view of security concerns; or should the main objective of their research be compromised by making data openly accessible.
The Pilot will give the Commission a better understanding of what supporting infrastructure is needed and of the impact of limiting factors such as security, privacy or data protection or other reasons for projects opting out of sharing. It will also contribute insights in how best to create incentives for researchers to manage and share their research data.
The Pilot will be monitored throughout Horizon 2020 with a view to developing future Commission policy and EU research funding programmes.
The 6th Africa-EU Cooperation Forum on ICT took place on December 2-3 2013 at the African Union Conference Center in Addis Ababa, Ethiopia under the aegis of the European Commission and the African Union Commission.
The event was organised by the EU EuroAfrica-P8 project on the occasion of the 50th anniversary of the African Union among the African ICT week.
During 20 sessions, 300 participants had the opportunity to share knowledge and explore the possibilities for cooperation in the framework of the Joint Africa-EU Strategic Partnership (JAES).
This meeting offered opportunities to the pro-iBiosphere project to promote its activities through the dissemination of project brochures and to network with potential stakeholders from Africa such as representatives from the Ethiopian agriculture portal on the occasion of the session on ICT for Agriculture on December 3.

Data analysis is a complicated and time-consuming process. Like a craftsman, you require a set of tools that source, reformat, merge and analyse data. Using these tools manually in a workflow can take weeks. Then, when you finally get the workflow working, you often need to run it again with a new set of inputs and parameters. What if there were a piece of software that could couple all these tools together and then run it all over again at a click of a button?. This is what Taverna Workbench does. Taverna, changes a time-consuming job with multiple tools into a single machine that does all the work seamlessly.
Taverna Workbench is one of the tools that supports the BioVel project with their stated aim of creating a "virtual e-laboratory that supports research on biodiversity issues". Taverna, by itself, is like a conductor without an orchestra. The power of Taverna is in the flexible coupling together of web-services, scripts and all kinds of processing engines to create workflows. For example, an obvious use case is the coupling together of the webservice from GBIF with a niche model engine and rerunning of the workflow using different projections of future climate change. However, Taverna can be used to simplify the processing of practically any digital data. Many ecologists use R as their primary statistical software. R can be run from within Taverna, but Taverna helps you couple its running to pre- and post-processing so that it can be run more easily.
Taverna is widely used among phylogeneticists and bioinformaticians, but other disciplines are rapidly adopting it. Another, unique and powerful feature of Taverna is that people can share, distribute, and collaborate on their workflows. On the website myExperiment.org scientists post their workflows for others to use, critisise and improve upon. The website works like a social network, enabling users to create groups, "like" favorite workflows and exchange ideas. You could spend the time to program your own links between services, but Taverna lets you do this easily without sacrificing innovation and adaptability.
One of the important features of Taverna is the seamless way it allows users to use webservices. There is a growing list of webservices for biodiversity from organisations such as GBIF, EOL and EU-Brazil OpenBio. One of the big issues with webservices is that they are, almost by definition, invisible to human users. Therefore, how do you find out that they exist?. This is where biodiversitycatalogue.org comes in. It allows scientists to discover webservices, but also describes how they work and their input and output formats. The pro-iBiosphere project has helped to improve the catalogue and will help to set priorities for future development. It now recommends the use of the Biodiversity Catalogue as central service registration facility for a future Open Biodiversity Knowledge Management System.
Taverna is a relatively new system to the Biodiversity community and through the BioVeL project its user-base is growing rapidly. Furthermore, people are finding new uses for it all the time.
On 5th December 2013, BioVel organised a workshop on Taverna at which pro-iBiosphere was represented. One potential use-case that is of interest to pro-iBiosphere is in the automated markup of text. Some aspects of automated markup are common to many texts, such as the identification of scientific names. On the other-hand there are other aspects that are specific to particular texts, such as the identification of treatment boundaries and language specific features. Taverna may be used to link generic services with custom scripts to significantly reduce the time it takes to markup text. Workflows could be created for one particular publication and then tweaked to work for another.
The possibilities of Taverna are almost limitless. It is just the glue, and you decide what you stick together. You might think the Taverna sounds like a quiet place for a drink, whereas, it is really the factory floor of data processing.
By Quentin Groom (National Botanic Garden of Belgium)
A pro-iBiosphere workshop on evaluation of business models took place on the 10th of October 2013 at the Botanischer Garten und Botanisches Museum (FUB-BGBM) in Berlin, Germany. It was attended by project partners and four external experts. The workshop was split into two sessions, each divided into smaller working groups. In the first session, a prioritised list of the partners' current products and services was drawn up, and the opportunities for, and threats to these were assessed. In the second session, the participants focussed on the services and activities that would comprise a future OBKMS (Open Biodiversity Knowledge Management System) and documented the constraints that might prevent the projected benefits of OBKMS from being realised.
The sessions have been very fruitful in terms of content (more than 20 matrix were made) and all participants (including external participants) have been very active during the whole day. Having external participants represented a real asset as they helped shaping the project vision more precisely while also demonstrating and confirming their interest in the OBKMS. Partners found these exercises very productive while taking time to step back and envision the future of the Consortium all together. This workshop is not the end of the exercise but only a milestone to agree on the various concepts, methodology and tools to be used to envision project sustainability and allow discussions among the partners. All in all, workshop objectives have been achieved.
The next steps of this workshop will be the release of an event report detailing the event outputs in presenting the project exploitation potential. We will keep you updated on the development of these activities.
For complementary information on the workshop (concept & objectives, agenda, participants list and presentations), visit the dedicated wiki page here.

From Oct 8-10th 2013 the 4th pro-iBiosphere meeting took place at the Botanic Garden and Botanical Museum Berlin-Dahlem. In total, 87 participants from 15 countries attended the 4 workshops held on:
1. 8 Oct Workshop 1 (M4.1): How to improve technical cooperation and interoperability at the e-infrastructure level (FUB-BGBM). For results from the workshop, see here.
2. 8 Oct Workshop 2 (M4.2): How to promote and foster the development & adoption of common mark-up standards & interoperability between schemas (PLAZI)
3. 9 Oct Workshop 3 (M6.2.2): Workshop on user engagement and benefits (RBGK)
4. 10 Oct Workshop 4 (M6.3.2): Towards sustainability towards service: Meeting to evaluate business models currently in use by partners and relevant non-partners (SIGMA)
Networking
A questionnaire was sent to the participants of the meeting. A total of 87 persons answered the questionnaire. The meeting received an overall positive feedback. The agendas of the workshop and the possibility to network were the strongest attractions for attending. Half of the delegates were able to establish or strengthen 5-10 contacts during the event (see Figure 1). Delegates appreciated the discussions and welcoming atmosphere. Despite that, they mentioned that there could have been more time allocated for each workshop and each break for discussions.
Workshops
Participants expressed their preference to work in small groups with well-defined targets. The need for presentations was very low, provided that the workshops are well focused and give ample time for discussion.
30% of the participants judged the quality of the workshop as high, 31% as very good, 13% as acceptable and 1% as below expectations (see Figure 2). 50% of the participants are interested in attending other pro-iBiosphere events in the future and 64 out of 87 persons would recommend them to colleagues.
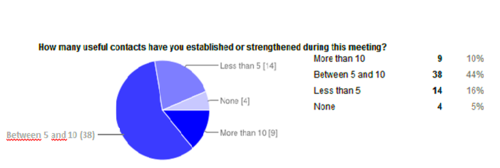
Caption: Contacts established by participants during the pro-iBiosphere meeting (first column; clockwise: 38% of the participants were able to make between 5 and 10 contacts, second column: 14% of the participants were able to establish less than 5 contacts, third column: 4% of the participants were not able to establish any contacts, fourth column: 9% of the participants were able to establish more than 10 contacts).
A pro-iBiosphere workshop on "How to improve technical cooperation and interoperability at the e-infrastructure level" was held at the Botanic Garden and Botanical Museum Berlin-Dahlem (BGBM) on October 8 2013. A total of 22 participants were invited to attend the workshop, representing a wide range of international biodiversity-related institutions and e-infrastructures. The workshop focused on the establishment of two highly relevant interoperability aspects of: (i) a consistent space of stable identifiers for collection objects across European taxonomic institutions; and (ii) a central registry for biodiversity-related services.
In the workshop 8 different implementations of stable http-URI-based identifier systems in European- and US-based taxonomic institutions where positively evaluated. These implementations are an important outcome of the fruitful collaboration between pro-iBiosphere and the Information Science and Technology Commission (ISTC) of the Consortium of European Taxonomic Facilities (CETAF).
In addition, the workshop conducted a thorough analysis of the BiodiversityCatalogue (https://www.biodiversitycatalogue.org/) developed by the University of Manchester in the context of the EU 7th Framework project BioVeL. As a result, a detailed list of recommended improvements of the Catalogue was compiled and agreed on. The University of Manchester will use these recommendations for setting priorities when further developing the Catalogue. Detailed results from the workshop are available here.
Authors: Anton Güntsch, Sabrina Eckert (FUB-BGBM)

The most visible forum for ICT research and innovation in Europe "ICT2013: Create, Connect and Grow", took place on the 6-8th of November 2013 in Vilnius (Lithuania). The event consisted of c.250 sessions and 200 exhibitors; and brought together lead thinkers and people driving European ICT research and innovation. A total of 6.000 persons participated in the event, including researchers, innovators, entrepreneurs, industry representatives, and politicians.
ICT 2013 allowed participants to share best practices and experiences in big data management, and provided them an excellent opportunity to learn about the current state of ICT research in Europe and the new Horizon2020 Framework programme for Research and Innovation.
The pro-iBiosphere project was strongly represented during the event by means of an exhibition booth and a networking session co-organised with other EC-funded projects (i.e. ei4Africa, Chain-reds, e-Science Talk). The exhibition booth entitled ‘e-Infrastructures at work and the future of research' showcased information from these four projects. Potential contacts were made with 20 stakeholders comprising projects on biodiversity data, EC-funded projects managing big data infrastructures (i.e. platforms, storage); and engineers specialised in semantic integration, enhancement, oncology, and autonomics.
The networking session on ‘What does the future hold for e-Science and Big Data?' brought together researchers, data owners and service providers (including SMEs) to explore the future for e-science and how to deliver open access to data through Horizon2020. During this session, the pro-iBiosphere project (represented by Plazi) presented its vision and potential impact to the biodiversity community and beyond. The networking session led to better understanding of how e-infrastructures can solve scientific challenges. Additional information is available here.
 Panel participants during the networking session ‘What does the future hold for e-Science and Big Data’
Panel participants during the networking session ‘What does the future hold for e-Science and Big Data’
Vibrant workshop for users of the EDIT Platform for Cybertaxonomy
The Vibrant workshop for users of the EDIT Platform for Cybertaxonomy was held from 11-13 November at the Botanic Garden and Botanical Museum Berlin-Dahlem (FUB-BGBM). The aim of the workshop was to explain the new Taxonomic Editor, the Data Portals and other components of the Platform to users, as well as to give an introduction to the software for structured taxonomic descriptions and keys, Xper2. The meeting brought together 40 participants representing a wide range of international biodiversity-related institutions and e-infrastructures (Euro+Med Plantbase, e-Floras, German Red Data Book editors, Pensoft publishers, Chinese Virtual Herbaria, Atlas Florae Europaeae and more).
The workshop was split into two parts, 1.5 days for the EDITor and the data portals, 0.5 days for Xper2. For the EDITor and data portal workshop, 3 parallel working groups (1 in German and 2 in English language) were led by two developers and one or two taxonomists from the FUB-BGBM. One group was solely formed by Euro+Med Plantbase family editors and taxonomic experts. For Xper2 two parallel groups were led by four colleagues from Paris. To explain the new Taxonomic EDITor two virtual box images where created (the images had the software - data Portal, Taxonomic Editor and cdmserver preinstalled). One image included a simple dataset with 10 taxa and factual data from the Compositae tribe Cichorieae. The other image included a more complex dataset from Euro+MedPlantBase, to give the editors the opportunity to do the hands-on training with the data they are handling in the framework of this project.
All three groups were able to get extensive hands-on experience with the Taxonomic EDITor and to observe the direct interaction of the EDITor with the data portals. Feedback from the users was generally positive. Some of them expressed the need of a "light" version of the EDITor which makes it easier to use for less experienced users. Constructive criticism from the participants will help to improve functionalities of the EDITor, and a part of them will use the EDIT platform in the future for their projects. The introduction and hands-on training on Xper2 was also well received by the audience and yielded some fruitful feedback for the presenters of that part of the workshop.
 Logo
Logo





 pro-iBiosphere wiki platform
pro-iBiosphere wiki platform
 RSS news
RSS news









