-
Step 1: Convert printed taxonomic articles/monographs to digital text format.
-
Step 2a: Mark up generic document features and domain-specific information (taxon treatments) and store the results at Plazi; and also
-
Step 2b: Export of newly published treatments marked up during the editorial process (for example in the journals ZooKeys, PhytoKeys and Mycokeys)
-
Step 3: Browse, search, export and re-use treatments coming from different sources.
-
Step 1. XML message from the publisher to the registry on acceptance of the manuscript containing the type of act, taxon names, and preliminary bibliographic metadata; the registry will store the data but not make these publicly available before the final publication date.
-
Step 2a. Response XML report containing the unique identified of the act as supplied by the registry and/or any relevant error messages.
-
Step 2b. Error correction and d-duplication performed manually: human intervention at either registry’s or publisher’s side (or at both).
-
Step 3. Inclusion of registry supplied identifiers in the published treatments (protologues, nomenclatural acts).
-
Step 4. Making the information in the registry publicly accessible upon publication, providing a link from the registry record to the artice.
- Steps forward 1: Implementation of HTTP-URIs by 8 major institutions for their collection objects by October 2013 and recommendations for further topics to be explored in detail.
- Steps forward 2: Agreement on the BiodiversityCatalogue as a global registry for biodiversity related services. Improvement recommendations for it to be able to fill this role even better, registration of services available now.
- Steps forward 3: Workflow improvement between the Plazi document registry and the Common Data Model (CDM)-based EDIT Platform for Cybertaxonomy (http://wiki.pro-ibiosphere.eu/wiki/Pilot_3 ). In the course of this a markup granularity table evolved. The pro-iBiosphere pilot portals visualize the data results at different stages and show the possibilities for scientists willing to mark up their data. The markup granularity table explains in detail work load and connected output gain.
-
To facilitate re-use and enhancement of biodiversity knowledge by a broad range of stakeholders, such as ecologists and niche modelers.
-
To foster a community of experts in biodiversity informatics and to build human links between research projects and institutions.
Italian National Research Center), Christian Brenninkmeijer (University of Manchester), Hannes Hettling (Naturalis Biodiversity Center), Rutger Vos (Naturalis Biodiversity Center)
[2] http://phenoscape.org/
[3] http://phenoscape.org/wiki/Phenex
[4] http://www.plantontology.org
[5] http://obofoundry.org/wiki/index.php/PATO:Main_Page
[6] Gkoutos, G. V., Green, E. C., Mallon, A.-M. M., Hancock, J. M., and Davidson, D. (2005) Using ontologies to describe mouse phenotypes. Genome biology, 6(1).
[7] Mungall, C., Gkoutos, G., Smith, C., Haendel, M., Lewis, S., and Ashburner, M. (2010) Integrating phenotype ontologies across multiple species. Genome Biology, 11(1), R2+.
[8] Robinson, P. N. et al. (2008) The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. American journal of human genetics, 83(5), 610–615.
[9] http://www.jbiomedsem.com/content/4/1/43
-
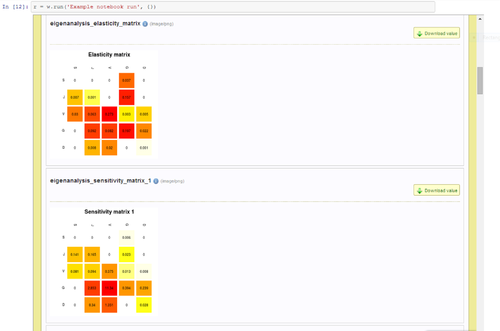
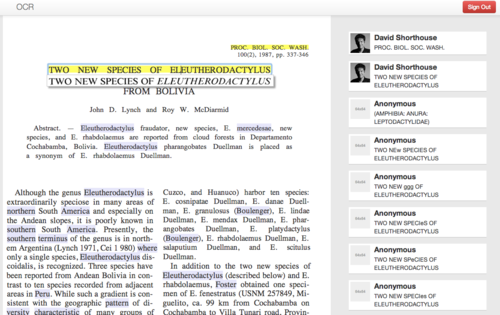
An in-place panel shows the exact line in the original scanned image while the user edits a single line of OCR text at a time (Figure 1)
-
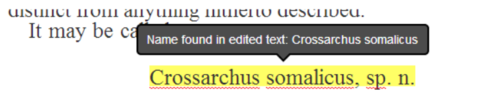
Global Names scientific name-finding is integrated in real-time when a user completes a line edit, giving feedback if a scientific name is newly recognized (Figure 2)
-
Authentication uses the facile https://oauth.io/ such that all edits are tied to users’ OAuth2-provider accounts (eg Google, Twitter, GitHub)
-
Frequencies of common edits are summarized in real-time and other words that may benefit from similar edits are highlighted for users
-
Batch processes collapse all user edits and text files are recreated for possible re-introduction into data mining routines
-
Unit and integration tests are included
The event wiki page here has been recently updated with additional information on the different series of activities organised (workshops, trainings and demonstrations) and the Final Conference agenda now comprises worldwide high-level speakers, including (i) officials from the European Commission DG Connect, the US National Academy of Sciences, (ii) representatives from botanic gardens, natural museums, other biodiversity initiatives and (iii) experts or (iv) researchers specialized in biodiversity informatics, environmental/natural science.
One of the key objectives of these series of events will be to ensure the Final event will provide key recommendations and inputs from biodiversity experts for the preparation of the next WP 2016-2017 as specifically asked by the European Commission.
The number of registered attendees has already reached a good level of participation to insure a thorough exchange of information and experience between stakeholders interested in making fundamental biodiversity data digital, open and re-usable. Visit the different activities pages to find out more on the attendance status.
In this context, if you plan to attend and have not yet registered, we can only recommend you do to it as soon as possible here (due to room capacity constraints).
For further information on this event (agenda, concept & objectives, registration) please visit the Event wiki page or contact us at [email protected].

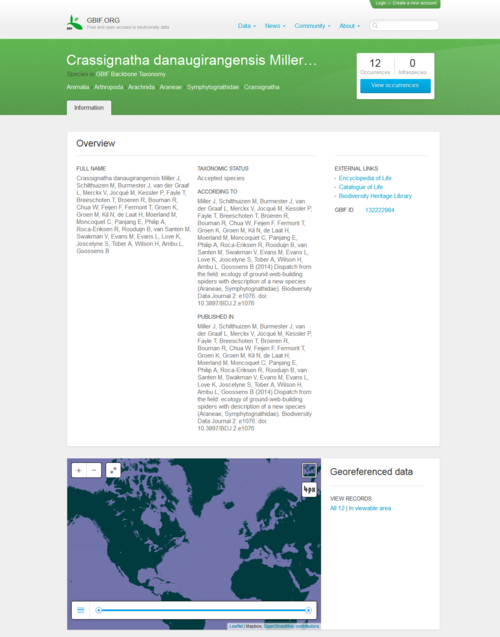 A group of scientists and students discovered the new species of spider during a field course in Borneo, supervised by Jeremy Miller and Menno Schilthuizen from the Naturalis Biodiversity Center, based in Leiden, the Netherlands. The species was described and submitted online from the field to the Biodiversity Data Journal through a satellite internet connection, along with the underlying data . The manuscript was peer-reviewed and published within two weeks of submission. On the day of publication, GBIF and EOL have harvested and included the data in their respective platforms.
A group of scientists and students discovered the new species of spider during a field course in Borneo, supervised by Jeremy Miller and Menno Schilthuizen from the Naturalis Biodiversity Center, based in Leiden, the Netherlands. The species was described and submitted online from the field to the Biodiversity Data Journal through a satellite internet connection, along with the underlying data . The manuscript was peer-reviewed and published within two weeks of submission. On the day of publication, GBIF and EOL have harvested and included the data in their respective platforms.
Workshop on Model Evaluation
Wednesday June 11 (all day)
Demonstrations on pro-iBiosphere pilots
Demonstrations on outcomes of pro-iBiosphere Data Enrichment Hackathon
Workshop on Biodiversity Catalogue
Training on WikiMedia
Poster session
Thursday June 12 (all day)
Final Conference
Networking Cocktail
 piB Final Event announcement
piB Final Event announcement
The iMarine initiative provides a data infrastructure aimed at facilitating open access, the sharing of data, collaborative analysis, processing and mining processing, as well as the dissemination of newly generated knowledge. The iMarine data infrastructure is developed to support decision making in high-level challenges that require policy decisions typical of the ecosystem approach.
iMarine has developed a series of applications which can be clustered in four main thematic domains (the so called Application Bundles, set of services and technologies grouped according to a family of related tasks for achieving a common objective).
More information on the iMarine Catalogue of Applications here.

BioVeL announces the upcoming release of the BioVeL Portal. Designed in response to scientists' needs through a continuous cycle of requests and feedback, the portal will be robust and scalable for handling greater workloads.
An important feature of the Portal will be the ability to do "data sweeps"– that is, to initiate multiple runs of the same workflow, each with different input conditions. Other neat points are the organisation of the workflows by categories with "facetted browsing" for easier search, and a complete history of all your own workflow runs. Also through the Portal you can share workflows and results between collaborators. And as always with BioVeL tools, the codebase used for the portal benefits from being used across multiple projects.
Access BioVeL Portal here.
Please do not hesitate to provide your comments to [email protected]

The pro-iBiosphere project organied 2 workshops between February 10-12 in Berlin:
- February 10: MS12 - Workshop on mark-up of biodiversity literature
- February 11: MS12 - Workshop on mark-up of biodiversity literature and MS23 - Workshop on alternative business models
- February 12: MS23 - Workshop on alternative business models
The workshops took place at the Museum für Naturkunde Berlin (MFN) located 43 Invalidenstraße in Berlin.
For complementary information on these events (concept, objectives and outcomes), please visit the dedicated project wiki page here or contact us: [email protected].






 pro-iBiosphere wiki platform
pro-iBiosphere wiki platform
 RSS news
RSS news





